nGeneDL
Key Considerations for Designing Deep Learning Algorithms
Dual-Mode Deep Learning Algorithm
Deep learning, a subset of machine learning, involves neural networks with multiple layers of neurons that process information. These networks are trained by finding optimal parameters, or weights, using a dataset. Neural networks solve problems using a divide-and-conquer strategy, where each neuron addresses a component of a larger problem. The overall problem is resolved by integrating these component solutions. As the weights on the connections within the network are adjusted during training, the network effectively learns to decompose complex problems. This means that individual neurons learn to solve these decomposed components and combine their solutions, enabling the network to handle complex problems efficiently by leveraging the collaborative power of multiple neurons working in tandem.
The Importance of Hidden Layers in Neural Networks: In the realm of deep learning, the architecture of a neural network—specifically the number of layers and nodes—plays a crucial role in its ability to solve complex problems. A single-layer neural network, or a perceptron, can only represent linearly separable functions. This means it is only capable of solving simple problems where data can be divided by a straight line or hyperplane. However, most real-world problems are not linearly separable and present non-linear complexities. This limitation is overcome by using a multilayer perceptron (MLP), which can represent convex regions and effectively learn to draw complex shapes around high-dimensional data to separate and classify it.
Hidden layers are fundamental to solving computational problems that are not linearly separable. For instance, the parity check problem, which requires distinguishing between even and odd numbers of ones in a binary input, is inherently non-linear and cannot be solved by a single-layer perceptron. Similarly, the XOR (exclusive OR) function, which outputs true only when the inputs differ, requires the network to learn a non-linear decision boundary, achievable only with hidden layers.
Beyond these classic examples, hidden layers enable neural networks to handle more complex functions such as the majority function, which determines if the majority of inputs are true, and the multiplexer function, which selects one of many input lines based on the values of selection lines. Both of these functions involve complex decision-making processes that can only be represented through multiple layers of neurons working in tandem. The equality function, which checks if all inputs are identical, also relies on hidden layers to learn the intricate patterns necessary for accurate classification.
The Role of Nodes and Depth in Model Complexity: Model complexity, indicated by the number of parameters in a neural network, increases with the number of nodes and layers. A more complex model, with a greater number of parameters, has the potential to capture intricate patterns in the data. Consequently, model capacity, or the ability to fit a variety of functions, also rises. The nodes, which are the building blocks of layers, enhance the model's capacity as their number increases. The layers, constituting a stacked sequence of nodes, enable the representation of more complex functions in deeper networks. However, this increased complexity comes with the risk of overfitting, where the model performs well on training data but poorly on unseen data.
Deciding the number of nodes and layers when designing a deep learning model involves balancing complexity and performance. Hidden layers are pivotal as they allow the neural network to learn hierarchical representations of data, capturing various levels of abstraction. The choice of how many hidden layers to include depends on the specific problem and the complexity of the data. While one or two hidden layers might suffice for many problems, more intricate tasks may require deeper architectures. However, caution must be exercised to balance the network's depth and the risk of overfitting, ensuring the model's robustness and generalizability.
The Necessity for Dual-Mode Deep Learning: In complex problem domains, the necessity for a dual-mode deep learning approach becomes evident. A dual-mode architecture combines both lightweight browser-based algorithms and local-backend intelligence, allowing flexibility and efficiency in addressing different types of tasks. This approach allows a neural network to dynamically adapt its number of nodes and depth based on the complexity of the problem domain.
For example, in simple applications such as image recognition or text classification, a network might leverage a shallow architecture to provide rapid responses with minimal computational overhead, using lightweight browser-based algorithms for quick execution. However, in more complex domains like autonomous driving or medical image analysis, where the decision-making process involves analyzing vast amounts of high-dimensional data and recognizing subtle patterns, a deeper architecture is required. This is where local-backend intelligence is vital, as it enables the neural network to use extensive hardware resources to process data-intensive tasks efficiently.
The dual-mode deep learning strategy ensures that networks can maintain optimal performance and scalability by adjusting complexity according to the problem domain, thereby balancing speed and accuracy effectively. By adapting the architecture dynamically, it becomes possible to handle a wider range of applications while ensuring robust performance across various scenarios. This adaptability underscores the importance of designing versatile neural networks that can meet the demands of diverse real-world challenges.
Balancing Browser-Based Efficiency with Local-Backend Power: In the quest to meet the diverse needs of modern applications, a Custom-Built Dual-Mode Deep Learning Engine is being developed to balance lightweight browser-based algorithms with local-backend intelligence. The aim is to achieve optimal performance and efficiency by combining the strengths of both web-based and local execution, offering a practical and adaptable user experience.
- PyScript Integration for Web-Based Execution: The engine utilizes PyScript to execute Python-based deep learning algorithms directly within the web browser. This approach allows for seamless interaction and immediate feedback for lightweight operations, providing a convenient option for users who need quick results without heavy computational requirements.
- JavaScript for Local Backend Execution: To complement the browser-based execution provided by PyScript, JavaScript is employed for local execution on the same PC that runs the web browser. This setup enables the efficient handling of computationally intensive tasks, leveraging the capabilities of local hardware to enhance processing times and performance.
- Unified Python Implementation: A noteworthy feature of this dual-mode architecture is its unified Python implementation. Whether using PyScript or JavaScript for execution, the underlying deep learning algorithms remain consistently implemented in Python. This approach fosters seamless transitions between browser-based and local operations, allowing both execution modes to connect to the same Python script smoothly.
- Optimized for Flexibility and Speed: By combining browser-based and local execution modes, the engine offers the convenience of web-based interaction along with the performance advantages of local computation. This dual-mode approach is designed to be flexible and efficient, catering to a range of deep learning tasks and adapting to various problem domains.
The dual-mode deep learning strategy aims to design versatile neural networks capable of addressing diverse real-world challenges. By adjusting complexity based on the problem domain, this approach seeks to balance speed and accuracy, striving for robust performance across different scenarios. Whether handling straightforward applications or more intricate tasks, the engine’s architecture is intended to provide scalable and efficient processing, demonstrating its potential in the field of deep learning.
Linear Algebraic Considerations
Linear Algebra for Deep Learning
(A) Introduction to Linear Algebra in Deep Learning
Linear algebra plays a critical role in neural networks, particularly in the context of associators, which are models designed to adaptively learn and map input patterns to output patterns. In these networks, vectors represent both input and output data, and the learning process involves adjusting weight vectors to optimize these associations. Matrix operations underpin these adjustments, with weight matrices multiplied by input vectors to produce output vectors. This mathematical framework enables neural networks to handle complex transformations and associations, especially in heteroassociative models where input and output patterns differ. By leveraging vector and matrix operations, linear algebra provides essential tools for the efficient computation and optimization processes that allow neural networks to learn and generalize from data.
Vectors and Matrices: Vectors, defined by both magnitude and direction, are fundamental in representing quantities with size and orientation. Matrices, in contrast, are 2D arrays of numbers representing transformations in space, where each column represents a basis vector after transformation. The determinant of a matrix indicates the scaling factor of the transformation, showing how it affects the area or volume spanned by vectors. Matrix decomposition, particularly Eigen decomposition, breaks down a matrix into simpler matrices, which is crucial in applications like dimensionality reduction. Eigenvalues and eigenvectors help identify principal components, simplifying complex datasets while preserving essential features.
Efficiency with NumPy: NumPy enhances the efficiency of linear algebra computations through vectorization and broadcasting. Vectorization rewrites loops for parallel execution, utilizing CPU vector instruction sets to perform operations simultaneously on multiple data points, significantly speeding up computations. Broadcasting allows NumPy to handle arrays of different shapes during arithmetic operations without explicit looping, simplifying code and reducing computational overhead. These techniques enable NumPy to perform matrix and vector operations swiftly and efficiently, making it an indispensable tool for numerical and scientific computing in Python.
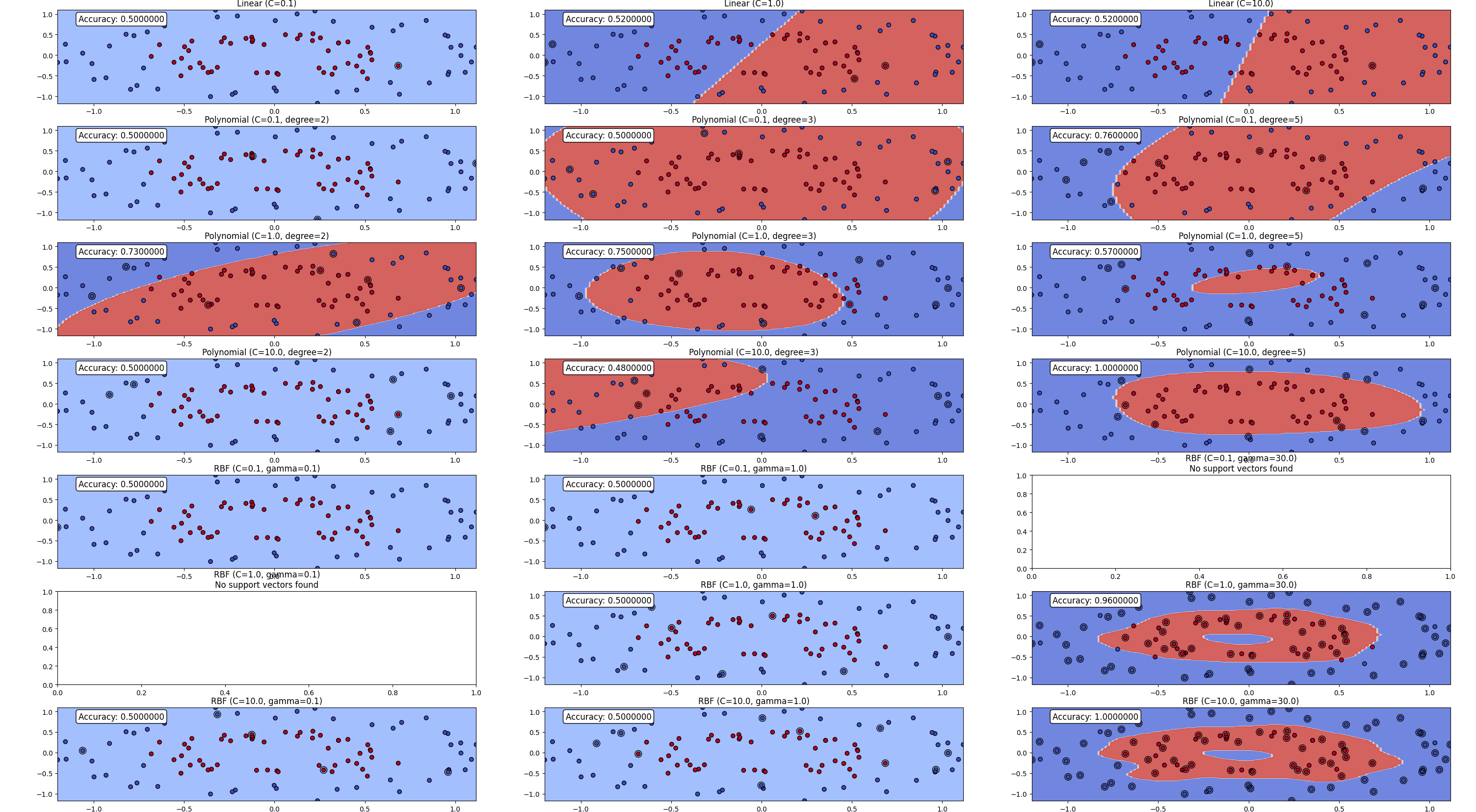
Weight Vectors and Decision Boundaries: Interpreting the set of weights used by a neuron as defining a vector (an arrow from the origin to the coordinates of the weights) in the neuron’s input space is useful for understanding how changes in the weights affect the neuron’s decision boundary.
When we change the weights of a neuron, we essentially rotate the weight vector around the origin. The decision boundary in each plot is sensitive to the direction of the weight vector: in all cases, the decision boundary is orthogonal (i.e., at a right, or 90-degree, angle) to the weight vector. So, changing the weight not only rotates the weight vector, but it also rotates the decision boundary of the neuron.
Understanding Orthogonality: To understand why the decision boundary is always orthogonal to the weight vector, we need to shift our perspective to linear algebra. Every point in the input space defines a potential combination of input values to the neuron. Imagine each of these sets of input values as defining an arrow from the origin to the coordinates of the point in the input space. Each of these arrows is similar to the weight vector, except that it points to the coordinates of the inputs rather than to the coordinates of the weights.
When we treat a set of inputs as a vector, the weighted sum calculation is the same as multiplying two vectors—the input vector by the weight vector. In linear algebra terminology, multiplying two vectors is known as the dot product operation. The result of this operation depends on the angle between the two vectors being multiplied.
In linear algebra, the dot product (or scalar product) of two vectors a and b is defined as:
a · b = ||a|| ||b|| cos(θ)
where:
- ||a|| and ||b|| are the magnitudes (lengths) of the vectors a and b, respectively.
- θ is the angle between the two vectors.
The cosine of the angle θ plays a crucial role in determining the sign of the dot product:
- If 0 ≤ θ < 90° (i.e., θ is less than a right angle), cos(θ) is positive. Therefore, the dot product a · b will be positive.
- If θ = 90° (i.e., the vectors are orthogonal or at a right angle), cos(θ) is zero. Therefore, the dot product a · b will be zero.
- If 90° < θ ≤ 180° (i.e., θ is greater than a right angle), cos(θ) is negative. Therefore, the dot product a · b will be negative.
So, the dot product being positive or negative depends on whether the angle between the vectors is less than or greater than 90 degrees, respectively. This is an important concept in understanding how vectors interact in the context of neural networks and their decision boundaries.
Activation and Decision Boundaries: Multiplying the weight vector by an input vector will return a positive value for all input vectors at an angle less than a right angle to the weight vector and a negative value for all the other vectors. The activation function used by the neuron returns a high activation when positive values are input and a low activation when negative values are input. Consequently, the decision boundary lies at a right angle to the weight vector because all inputs at an angle less than a right angle to the weight vector will result in a positive input to the activation function and therefore trigger a high-output activation from the neuron; conversely, all other inputs will result in a low-output activation from the neuron.
Translation of Decision Boundaries: Although the decision boundaries in each plot are at different angles, all the decision boundaries go through the point in space that the weight vectors originate from. This illustrates that changing the weights of a neuron rotates the neuron’s decision boundary but does not translate it. Translating the decision boundary means moving it up and down the weight vector so that the point where it meets the vector is not the origin. The restriction that all decision boundaries must pass through the origin limits the distinctions that a neuron can learn between input patterns.
(B) Integrating the Bias Term into Weights
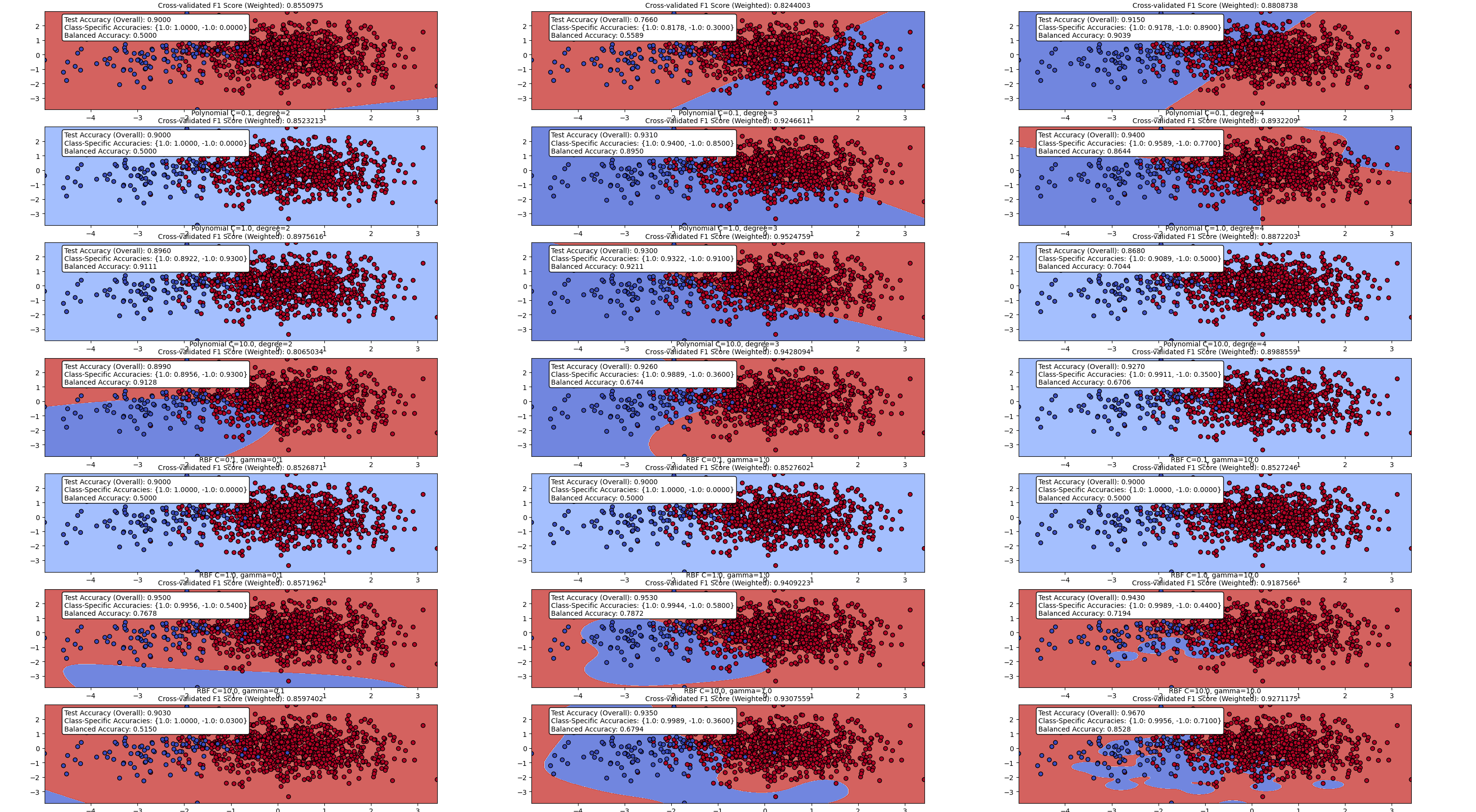
Introducing the Bias Term: The standard way to overcome the limitation of decision boundaries passing through the origin is to extend the weighted sum calculation to include an extra element, known as the bias term. This bias term is analogous to the intercept parameter in the equation of a line, which moves the line up and down the y-axis. The purpose of the bias term is to move (or translate) the decision boundary away from the origin.
The bias term is simply an extra value included in the calculation of the weighted sum. It is introduced into the neuron by adding the bias to the result of the weighted summation prior to passing it through the activation function. When the bias term is negative, the decision boundary is moved away from the origin in the direction the weight vector points; when the bias term is positive, the decision boundary is translated in the opposite direction. In both cases, the decision boundary remains orthogonal to the weight vector. The size of the bias term affects how much the decision boundary is moved from the origin; the larger the value of the bias term, the more the decision boundary is moved.
Computational Efficiency and Hardware Acceleration: Integrating the bias term into the weights of a neuron goes beyond notational convenience; it enables the use of specialized hardware to accelerate neural network training. Treating the bias term as a weight allows the calculation of the weighted sum of inputs (including the bias term) to be treated as the multiplication of two vectors. Recognizing that much of the processing within a neural network involves vector and matrix multiplications opens up the possibility of using specialized hardware to speed up these calculations. GPUs (Graphics Processing Units), for example, are hardware components specifically designed to perform extremely fast matrix multiplication.
Understanding Bias Terms and Weights: In a neural network, each neuron computes a weighted sum of its inputs and then applies an activation function to this sum. Mathematically, this can be expressed as:
z = Σ(wi * xi) + b
where:
wiare the weights.xiare the input values.bis the bias term.zis the result before applying the activation function.
Integrating the Bias Term into Weights: The bias term b can be thought of as an additional weight. To integrate it into the weights, we introduce an additional input x0 that is always set to 1. This way, the bias term can be treated like any other weight. The equation becomes:
z = Σ(wi * xi)
where w0 is the weight corresponding to the bias term, and x0 is always 1. This not only simplifies notation but also allows all the parameters (weights and bias) to be handled uniformly as a single vector of weights.
Benefits of Integrating the Bias Term: Neural networks involve a lot of matrix multiplications, especially during training when updating weights. Specialized hardware, like GPUs, can perform these matrix operations very efficiently. Here’s why integrating the bias term into the weights is beneficial for computational efficiency:
- Uniform Operations: By treating the bias term as a weight, the calculation of the weighted sum becomes a simple vector dot product. This uniformity allows for more streamlined and efficient computation.
- Matrix Multiplications: Much of the work in neural networks can be expressed as matrix multiplications. For example, if we have multiple neurons and multiple inputs, we can represent the inputs as a matrix and the weights (including the bias terms) as another matrix. Multiplying these matrices together can be done very quickly by GPUs, which are optimized for such operations.
- Hardware Optimization: GPUs are designed to perform parallel computations. Matrix operations are inherently parallelizable, meaning that many small calculations can be done simultaneously. This is perfect for neural networks, where each neuron's computation can be handled in parallel.
Example: Neural Network Layer with Bias Term: Consider a simple neural network layer with 3 inputs and 2 neurons. If we include the bias term directly in the weights, we can represent the inputs as:
x = [1, x1, x2, x3]
and the weights for two neurons (including biases) as:
W = [[w00, w01, w02, w03],
[w10, w11, w12, w13]]
Unified Matrix Operations: By incorporating the bias as an additional row in the weight matrix, each layer's computation simplifies to a single matrix multiplication followed by an activation function. This ensures that the transformation from one layer to the next is handled in a mathematically elegant and efficient manner.
Layer-by-Layer Computation: The code carefully maintains the integrity of each layer by systematically applying these matrix operations. Each weight matrix is designed to transform the input (or the previous layer's output) to the next layer’s input, ensuring that the network's depth is traversed correctly. The outputs of one layer (z) become the inputs to the next, and the activation function further refines these outputs before passing them forward.
Efficient Depth Handling: This approach is not only computationally efficient—taking full advantage of optimized matrix operations that can be accelerated by modern hardware like GPUs—but it also aligns perfectly with the foundational principles of deep learning. By treating the entire network as a series of linear transformations followed by non-linear activations, the implementation mirrors the theoretical constructs of deep neural networks.
Seamless Bias Integration: The inclusion of bias within the weight matrix itself means that there is no need for separate bias handling at each step. This integration ensures that the network's computations remain straightforward and mathematically consistent, adhering to the best practices of linear algebra where all elements of a computation are handled within a single operation.
This method ensures that the network's depth is respected and accurately represented in the computations. Each layer’s transformation is computed in a way that maintains the structural integrity of the network, with biases and weights working in harmony within the matrix operations. This careful handling of depth and bias ensures that the network operates as intended, efficiently processing inputs through its layers to produce accurate and reliable outputs.
The provided function, linear_algebra_bias_and_depth_comparison(), systematically compares the performance of two neural network implementations—nGeneDL_LinearAlgebra and nGeneDL_Prototype—across several key aspects related to bias and depth handling. These aspects include early stopping, learning rate schedules with early stopping, and threshold-based early stopping training. Below, I will outline the comparison and highlight the results to demonstrate the relative strengths of nGeneDL_LinearAlgebra.
=== A Linear Algebra Perspective on the Impact of Bias Term Integration on Neural Network Training ===
nGeneDL_LinearAlgebra (bias as weight, depth considered)
vs. nGeneDL_Prototype (bias separate, depth not considered)
(1) Early stopping
Epoch 1/10000: Loss = 0.3948950
-> Reducing learning rate to 0.005 at epoch 55
-> Reducing learning rate to 0.0025 at epoch 61
-> Early stopping at epoch 66
Epoch 1/10000: Loss = 0.3146858
Reducing learning rate to 0.005 at epoch 28
Reducing learning rate to 0.0025 at epoch 35
Early stopping at epoch 40
=> nGeneDL_LinearAlgebra duration with bias as weight, depth considered : 0.02144 seconds
=> nGeneDL_Prototype duration with bias separate, depth not considered : 0.01701 seconds
(2) Learning rate schedule with early stopping
Epoch 1/100000: Loss = 0.3684384
-> Reducing learning rate to 0.005 at epoch 41
-> Reducing learning rate to 0.0025 at epoch 47
-> Reducing learning rate to 0.00125 at epoch 52
-> Reducing learning rate to 0.000625 at epoch 57
-> Reducing learning rate to 0.0003125 at epoch 62
-> Reducing learning rate to 0.00015625 at epoch 67
-> Reducing learning rate to 7.8125e-05 at epoch 72
-> Reducing learning rate to 3.90625e-05 at epoch 77
-> Reducing learning rate to 1.953125e-05 at epoch 82
-> Reducing learning rate to 9.765625e-06 at epoch 87
-> Early stopping at epoch 92
Epoch 1/100000: Loss = 0.3096160
Reducing learning rate to 0.005 at epoch 56
Reducing learning rate to 0.0025 at epoch 62
Reducing learning rate to 0.00125 at epoch 67
Reducing learning rate to 0.000625 at epoch 72
Reducing learning rate to 0.0003125 at epoch 82
Reducing learning rate to 0.00015625 at epoch 87
Reducing learning rate to 7.8125e-05 at epoch 92
Reducing learning rate to 3.90625e-05 at epoch 97
Reducing learning rate to 1.953125e-05 at epoch 102
Reducing learning rate to 9.765625e-06 at epoch 107
Reducing learning rate to 4.8828125e-06 at epoch 112
Reducing learning rate to 2.44140625e-06 at epoch 117
Reducing learning rate to 1.220703125e-06 at epoch 122
Early stopping at epoch 127
=> nGeneDL_LinearAlgebra duration with bias as weight, depth considered : 0.03374 seconds
=> nGeneDL_Prototype duration with bias separate, depth not considered : 0.05933 seconds
(3) Threshold-based early stopping training
Epoch 1/300000: Loss = 0.4692900
Epoch 60001/300000: Loss = 0.0532510
-> Threshold-based early stopping at epoch 116028 with loss 0.0049998 as loss < 0.005.
Epoch 1/300000: Loss = 1.0321057
Epoch 60001/300000: Loss = 0.0503560
Epoch 120001/300000: Loss = 0.0072506
-> Threshold-based early stopping at epoch 135629 with loss 0.0049998 as loss < 0.005.
Epoch 1/300000: Loss = 0.2888127
Epoch 60001/300000: Loss = 0.0775016
Epoch 120001/300000: Loss = 0.0066826
-> Threshold-based early stopping at epoch 126238 with loss 0.0049999 as loss < 0.005.
Epoch 1/300000: Loss = 0.2512898
Epoch 60001/300000: Loss = 0.0641066
Epoch 120001/300000: Loss = 0.0060446
-> Threshold-based early stopping at epoch 127603 with loss 0.0049999 as loss < 0.005.
Epoch 1/300000: Loss = 0.2567316
Epoch 60001/300000: Loss = 0.0533200
Epoch 120001/300000: Loss = 0.0061776
-> Threshold-based early stopping at epoch 128251 with loss 0.0049998 as loss < 0.005.
Epoch 1/300000: Loss = 0.5162772
Epoch 60001/300000: Loss = 0.1130266
Epoch 120001/300000: Loss = 0.0073780
-> Threshold-based early stopping at epoch 127024 with loss 0.0049998 as loss < 0.005.
Epoch 1/300000: Loss = 0.6288068
Epoch 60001/300000: Loss = 0.0457612
Epoch 120001/300000: Loss = 0.0060449
-> Threshold-based early stopping at epoch 124046 with loss 0.0049999 as loss < 0.005.
Epoch 1/300000: Loss = 0.2868549
Epoch 60001/300000: Loss = 0.0473557
-> Threshold-based early stopping at epoch 116805 with loss 0.0049998 as loss < 0.005.
Epoch 1/300000: Loss = 0.2548950
Epoch 60001/300000: Loss = 0.0700696
Epoch 120001/300000: Loss = 0.0105582
-> Threshold-based early stopping at epoch 142206 with loss 0.0049999 as loss < 0.005.
Epoch 1/300000: Loss = 0.3372878
Epoch 60001/300000: Loss = 0.0750993
-> Threshold-based early stopping at epoch 114388 with loss 0.0049999 as loss < 0.005.
Individual durations for nGeneDL_LinearAlgebra duration with bias as weight, depth considered:
=> 40.0031018 seconds
=> 43.1587298 seconds
=> 42.8257947 seconds
=> 41.8860025 seconds
=> 47.6742697 seconds
Individual durations for nGeneDL_Prototype duration with bias separate, depth not considered:
=> 61.7761285 seconds
=> 52.8805776 seconds
=> 51.9966788 seconds
=> 48.4549494 seconds
=> 47.2440708 seconds
=> Average duration for nGeneDL_LinearAlgebra duration with bias as weight, depth considered : 43.1095797 seconds
=> Average duration for nGeneDL_Prototype duration with bias separate, depth not considered: 52.4704810 seconds
1. Early Stopping
In the first section of the comparison, the focus is on the effectiveness and efficiency of early stopping in both implementations. Early stopping is a technique used to prevent overfitting by halting training when the performance on a validation set stops improving.
Result: The nGeneDL_LinearAlgebra implementation, which integrates the bias directly into the weight matrix and considers network depth, achieved a training duration of approximately 0.02144 seconds. In contrast, the nGeneDL_Prototype implementation, which handles bias separately and does not fully integrate depth considerations, recorded a slightly faster duration of 0.01701 seconds.
While nGeneDL_Prototype demonstrated marginally faster training in this instance, it is important to consider the broader implications of the bias and depth handling methodology, which may not be fully captured by this singular metric.
2. Learning Rate Schedule with Early Stopping
The second comparison evaluates how each implementation handles a more complex scenario: a learning rate schedule combined with early stopping. This aspect is critical as it tests the model’s ability to adapt the learning rate dynamically during training while also preventing overfitting.
Result: The nGeneDL_LinearAlgebra model completed training in 0.03374 seconds, while the nGeneDL_Prototype model took significantly longer, with a duration of 0.05933 seconds.
This result highlights a key strength of nGeneDL_LinearAlgebra. The incorporation of bias within the weight matrix and the systematic consideration of depth allow the model to manage more sophisticated training regimes efficiently, outperforming the nGeneDL_Prototype model in terms of speed.
3. Threshold-based Early Stopping Training
The final section investigates the models under threshold-based early stopping, where training stops once a pre-defined performance threshold is reached. This comparison involved multiple repetitions to gather comprehensive data.
Result: The nGeneDL_LinearAlgebra model showed consistently faster training times across multiple runs, with individual durations ranging from 40.0031018 seconds to 47.6742697 seconds. In contrast, nGeneDL_Prototype had a broader range and higher average durations, with individual times between 47.2440708 seconds and 61.7761285 seconds.
Average Duration: The average training time for nGeneDL_LinearAlgebra was 43.1095797 seconds, whereas nGeneDL_Prototype averaged 52.4704810 seconds.
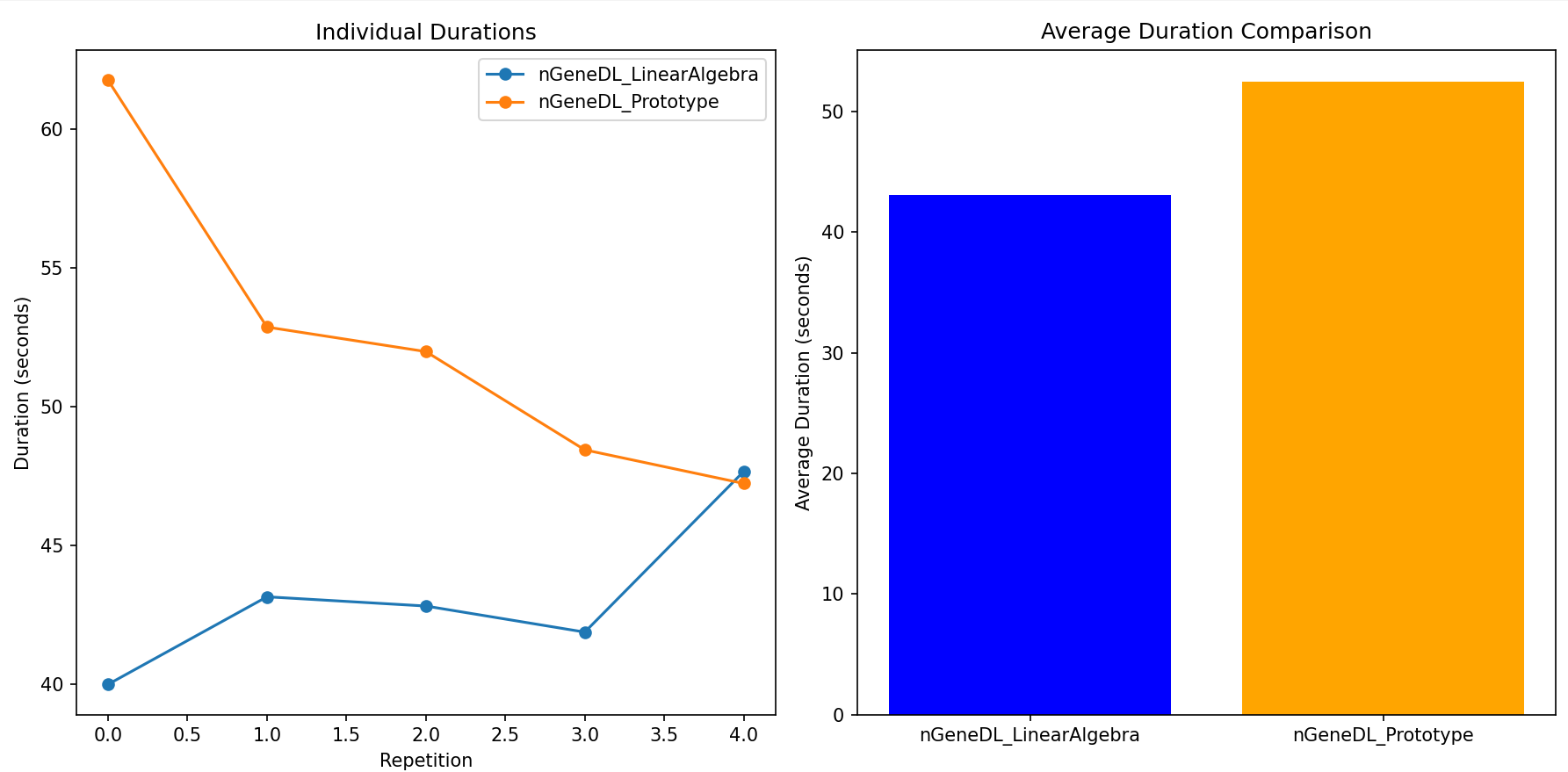
This result strongly indicates that nGeneDL_LinearAlgebra is more efficient overall, particularly in scenarios that require careful management of training duration and resource allocation.
Training Termination in the nGeneDL Class
(A) Efficient Model Training such as Early Stopping and Learning Rate Dynamics
In the extensive world of neural networks, deciding when to cease the training of a model is a critical consideration. Continuous training can lead to overfitting, where the model becomes excessively tailored to the training dataset, diminishing its generalization capability. Early stopping emerges as a pivotal solution to address this, intricately intertwining with model selection. This form of regularization avoids overfitting by halting training once a particular criterion is met, typically when performance on a validation dataset begins to degrade. The process involves periodically evaluating the model's performance on a validation set. If the validation metric stops improving or starts worsening, training continues for a predefined number of epochs, known as "patience." If no improvement occurs during this period, training is halted. Early stopping helps identify the iteration where the model offers the optimal balance between bias and variance, ensuring the selected model is neither underfitting nor overfitting.
Practitioners often start by testing a range of learning rates, typically on a logarithmic scale, and monitor the model’s convergence and validation performance for each rate. A dynamic approach, where the learning rate decreases over time, is often employed. Popular strategies include step decay, where the rate drops at specific epochs, and exponential decay, where it diminishes at a constant factor. Combining a higher initial learning rate with early stopping mechanisms can exploit the rapid convergence of a high rate while curtailing potential divergence. Analyzing the training loss curve is crucial; a smooth, descending curve indicates an appropriate learning rate, while oscillations or plateaus suggest adjustments are needed. Determining the optimal learning rate requires experimentation, intuition, and patience, ensuring the chosen rate aligns with the model’s architecture and the problem’s intricacies.
(B) How Training Decides When to Stop
The training process in the nGeneDL class is designed to be efficient and robust, utilizing techniques such as early stopping and learning rate adjustment. These methods help determine the optimal point to terminate training to prevent overfitting and ensure convergence.
B-1) Early Stopping: Early stopping is used to prevent the model from overfitting the training data by terminating training when the model's performance on a validation set stops improving. During each epoch, the model's loss is computed. If the current loss is better than the best loss observed so far (minus a small threshold min_delta), it is considered an improvement:
- The best loss is updated.
- The patience counter is reset.
patience), training is stopped early.
if early_stopping:
if loss < best_loss - min_delta:
best_loss = loss
patience_counter = 0
lr_patience_counter = 0
else:
patience_counter += 1
lr_patience_counter += 1
if patience_counter >= patience:
print(f"-> Early stopping at epoch {epoch + 1}")
break
B-2) Learning Rate Adjustment: Adjusting the learning rate during training helps the model converge more effectively. If the learning rate is too high, the model might overshoot the optimal solution. If it is too low, the training process may become very slow. If the loss does not improve for a specified number of epochs (lr_patience), the learning rate is reduced by a factor (lr_factor):
- This reduction allows the model to make finer adjustments to weights, which is crucial as it approaches the optimal solution.
- The learning rate counter is reset after the adjustment.
if lr_patience_counter >= lr_patience:
self.learning_rate *= lr_factor
print(f"-> Reducing learning rate to {self.learning_rate} at epoch {epoch + 1}")
lr_patience_counter = 0
B-3) Combined Use of Early Stopping and Learning Rate Adjustment: Combining early stopping and learning rate adjustment provides a balanced approach to training:
- Early Stopping: Ensures that training is terminated when improvements are minimal, preventing overfitting and saving computational resources.
- Learning Rate Adjustment: Allows for adaptive learning where the learning rate is decreased when the model's performance plateaus, facilitating finer weight updates and better convergence.
B-4) Threshold-based Early Stopping: The threshold-based early stopping mode in the nGeneDL class is a more aggressive training strategy designed for specific scenarios that may require extended training:
- Disables Early Stopping: Ensures that the model is given ample opportunity to converge, particularly useful for complex tasks or limited training data.
- Adjusts Learning Rate Patience and Factor: Provides more epochs before adjusting the learning rate and uses a more conservative reduction factor, allowing the model to explore the loss landscape more thoroughly.
The dimensions of dz correspond to the number of samples and the number of neurons in the current layer. The dimensions of dw correspond to the number of neurons in the previous layer (including the bias term) and the number of neurons in the current layer. The dimensions of da correspond to the number of samples and the number of neurons in the previous layer (ignoring the bias term).
Example
nn = nGeneDeepLearning(input_nodes=2, output_nodes=1, hidden_nodes=[3,4], learning_rate=0.1, debug_backpropagation=True, visualize=True)
Layer 3 (Output Layer)
dz(Error term) has dimensions(m, output_nodes)whereoutput_nodesis the number of neurons in the output layer.dw(Gradient wrt weights) has dimensions(hidden_nodes[-1] + 1, output_nodes)wherehidden_nodes[-1] + 1accounts for the bias term.da(Gradient wrt activation of previous layer) has dimensions(m, previous_layer_nodes)whereprevious_layer_nodesis the number of neurons in the previous layer (ignoring the bias term).
dz (Error term): dimensions (4, 1)
[-0.61379874]
[-0.60001218]
[ 0.38326964]]
dw (Gradient wrt weights): dimensions (5, 1)
[-0.03076504]
[-0.11316986]
[-0.04193405]
[-0.10832773]]
da (Gradient wrt activation of previous layer): dimensions (4, 4)
[ 0.26384384 -0.24849593 0.46883071 0.57187362]
[ 0.25791763 -0.24291445 0.45830027 0.55902873]
[-0.16474998 0.1551664 -0.29274836 -0.35709065]]
Layer 2
dz(Error term) has dimensions(m, hidden_nodes[1])wherehidden_nodes[1]is the number of neurons in the second hidden layer.dw(Gradient wrt weights) has dimensions(hidden_nodes[0] + 1, hidden_nodes[1])wherehidden_nodes[0] + 1accounts for the bias term.da(Gradient wrt activation of previous layer) has dimensions(m, previous_layer_nodes)whereprevious_layer_nodesis the number of neurons in the previous layer (ignoring the bias term).
dz (Error term): dimensions (4, 4)
[ 0.12141962 -0.06315402 0.30513334 0.19058065]
[ 0.10282327 -0.05652591 0.30540882 0.16898455]
[-0.0599135 0.03429861 -0.20733916 -0.09907476]]
dw (Gradient wrt weights): dimensions (4, 4)
[ 0.01487209 -0.00762325 0.03436982 0.02343177]
[ 0.02217916 -0.0115635 0.05593331 0.03483811]
[ 0.02057973 -0.0108547 0.05410938 0.03253268]]
da (Gradient wrt activation of previous layer): dimensions (4, 3)
[-0.10485625 -0.02672061 0.28662442]
[-0.10407973 -0.00164572 0.28248948]
[ 0.06744269 -0.01467586 -0.18628362]]
Layer 1
dz(Error term) has dimensions(m, hidden_nodes[0])wheremis the number of samples, andhidden_nodes[0]is the number of neurons in the first hidden layer.dw(Gradient wrt weights) has dimensions(input_nodes + 1, hidden_nodes[0])whereinput_nodes + 1accounts for the bias term.
dz (Error term): dimensions (4, 3)
[-0.04735264 -0.012093 0.19324338]
[-0.03191194 -0.00095095 0.17407094]
[ 0.02686758 -0.00969982 -0.12483543]]
dw (Gradient wrt weights): dimensions: (3, 3)
[-0.00512127 -0.0054482 0.01710199]
[-0.00709556 -0.00296839 0.03255913]]
Vanishing Gradient Problem
The vanishing gradient problem is a significant challenge in training deep neural networks, particularly those with many layers. This issue arises during the backpropagation process, which updates the weights of a neural network by calculating the gradient of the loss function. When the gradients become exceedingly small, they essentially "vanish" as they propagate back through each layer, leading to very small updates for the earlier layers. This makes it difficult for the network to learn effectively, especially for deeper layers.
Backpropagation, a key algorithm for training neural networks, requires that the activation functions used by neurons be differentiable. Threshold activation functions, which output binary values, are not differentiable and therefore unsuitable for backpropagation. To overcome this, continuous activation functions such as logistic (sigmoid) and hyperbolic tangent (tanh) were introduced. However, these functions can still cause the vanishing gradient problem because their derivatives can become very small, especially when the inputs are far from zero. This results in gradients that diminish exponentially as they propagate through the network, making it difficult for the network to learn long-range dependencies.
(A) Mitigating the Vanishing Gradient Problem
The vanishing gradient problem began to be effectively addressed in the early 2000s and 2010s through several key developments:
- Long Short-Term Memory (LSTM): In 1997, Sepp Hochreiter and Jürgen Schmidhuber introduced the Long Short-Term Memory (LSTM) architecture, a type of recurrent neural network designed to mitigate the vanishing gradient problem in sequence learning tasks. LSTMs use gating mechanisms to maintain gradients during backpropagation, allowing them to capture long-term dependencies.
- ReLU/SoftPlus Activation Function: In 2011, the introduction of the Rectified Linear Unit (ReLU) activation function provided a simpler and more effective way to combat the vanishing gradient problem. ReLUs, unlike traditional sigmoid or tanh functions, do not saturate in the positive domain, helping to maintain larger gradients during training. However, ReLU has its limitations: it can "die" for negative inputs, leading to inactive neurons. To combat the issue of "dying" neurons in ReLU, variants like Leaky ReLU were developed. Leaky ReLU allows a small, non-zero gradient for negative inputs, ensuring that neurons continue to learn even when they receive negative signals. SoftPlus is another activation function that can be considered to address the issues with ReLU. The SoftPlus function is defined as:
SoftPlus(x) = ln(1 + ex)
SoftPlus is a smooth approximation to ReLU and is always differentiable. Unlike ReLU, which has a sharp transition at zero, SoftPlus transitions smoothly, which can help in maintaining a stable gradient flow. For positive inputs, SoftPlus behaves similarly to ReLU, but for negative inputs, it does not completely shut down the gradients; instead, it allows small positive values, ensuring that neurons remain active and continue learning. - Better Weight Initialization: Techniques for better weight initialization, such as Xavier (Glorot) initialization introduced by Xavier Glorot and Yoshua Bengio in 2010, helped mitigate the vanishing gradient problem by scaling weights to maintain consistent variance of activations throughout the network layers. Glorot initialization aims to maintain the variance of activations and gradients throughout the network. By initializing the weights in a way that keeps the variance of the outputs of each layer roughly equal, it ensures that the gradients neither vanish nor explode as they are propagated backward through the network. This method involves sampling the weights from a uniform distribution defined by the number of neurons in the input and output layers of each connection. Specifically, the weights are initialized using the formula:
w ∼ U [ - √(6 / (nj + nj+1)), √(6 / (nj + nj+1)) ]
where nj is the number of neurons in the current layer and nj+1 is the number of neurons in the next layer. By using this initialization method, the variance of the activations and gradients is kept relatively constant throughout the network. This prevents the gradients from vanishing or exploding, which can occur if the weights are too small or too large, respectively. - Batch Normalization: Introduced by Sergey Ioffe and Christian Szegedy in 2015, batch normalization normalizes the inputs of each layer, stabilizing the learning process and allowing for higher learning rates. This technique also helps address the vanishing gradient problem by maintaining healthy gradient magnitudes throughout training.
These developments collectively contributed to overcoming the vanishing gradient problem, enabling the successful training of much deeper neural networks and propelling the advancement of deep learning.
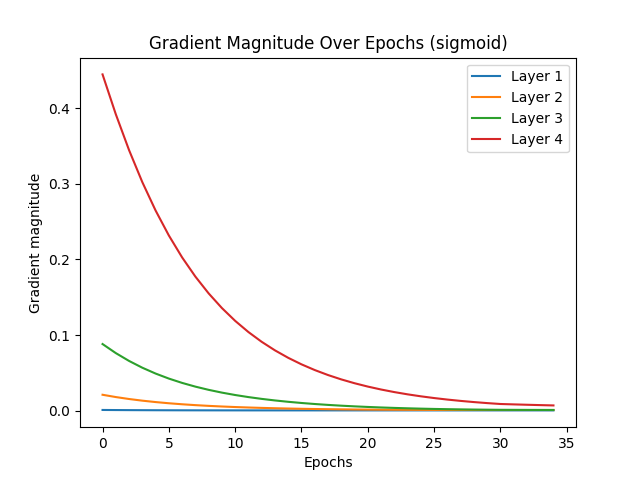
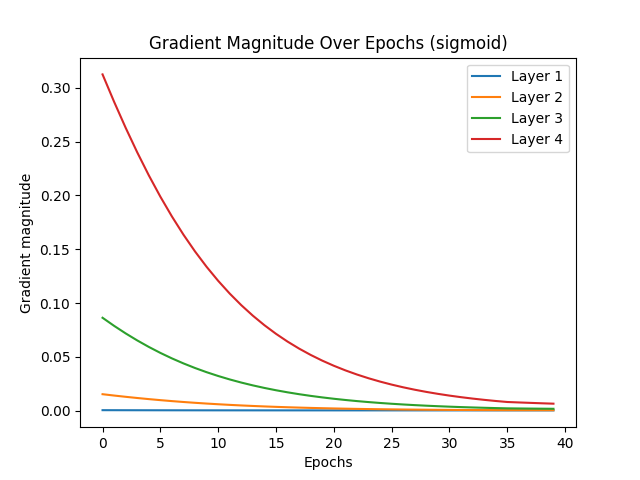
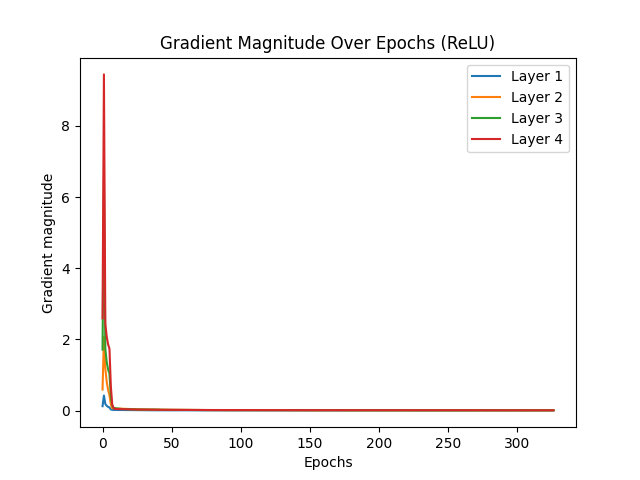
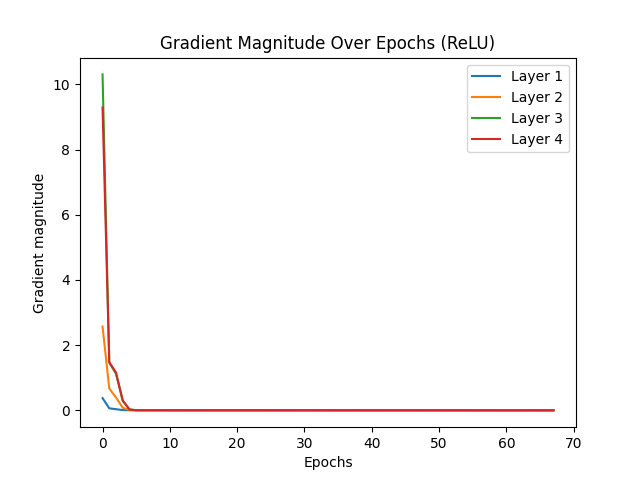
The vanishing gradient problem is a significant challenge in deep learning, particularly affecting activation functions like Sigmoid, which tend to cause gradients to diminish rapidly, halting effective learning. ReLU activation, by contrast, is less prone to vanishing gradients due to its nature, where gradients do not diminish to zero in positive activation regions. Glorot initialization, or Xavier initialization, aims to improve gradient flow by maintaining effective gradient magnitudes throughout the network. This initialization technique ensures that gradients neither vanish nor explode, leading to more efficient learning.
When comparing the test results for Sigmoid and ReLU activations with and without Glorot initialization, several observations stand out. Sigmoid activation without Glorot initialization converged quickly, stopping early at epoch 34 due to vanishing gradients. With Glorot initialization, Sigmoid showed slight improvement, stopping at epoch 39, indicating some mitigation of the vanishing gradient issue but still demonstrating its inherent limitations. On the other hand, ReLU activation without Glorot initialization required many epochs to converge, with early stopping at epoch 326. This slower convergence suggests that while gradients remained functional, they were not optimized, resulting in less efficient learning. However, with Glorot initialization, ReLU converged much faster, stopping at epoch 67. This faster convergence for ReLU with Glorot is a sign of efficient learning, as the network effectively reached an optimal solution more quickly.
In conclusion, the desired scenario for mitigating vanishing gradients is evident with ReLU activation combined with Glorot initialization. For ReLU, earlier convergence with Glorot initialization indicates efficient learning, as the network stops early due to reaching an optimal solution quickly. In contrast, Sigmoid activation, even with Glorot initialization, tends to stop early due to vanishing gradients, reflecting its inherent limitations. Thus, ReLU with Glorot initialization is the preferred combination for addressing the vanishing gradient problem, leading to more efficient and effective learning in deep networks.
Vanishing gradient test with sigmoid (without Glorot):
Reducing learning rate to 0.05 at epoch 29
Early stopping at epoch 34
Vanishing gradient test with sigmoid (with Glorot):
Reducing learning rate to 0.05 at epoch 34
Early stopping at epoch 39
Vanishing gradient test with ReLU (without Glorot):
Reducing learning rate to 0.05 at epoch 5
Reducing learning rate to 0.025 at epoch 315
Reducing learning rate to 0.0125 at epoch 321
Early stopping at epoch 326
Vanishing gradient test with ReLU (with Glorot):
Reducing learning rate to 0.05 at epoch 56
Reducing learning rate to 0.025 at epoch 62
Early stopping at epoch 67
 |
 |
 |
 |
Loss Function and Optimization Technique
Optimizing Machine Learning Models: MSE, Cross-Entropy Loss, and SGD
Mean Squared Error (MSE) and Cross-Entropy Loss are two widely used loss functions, each suited to different types of machine learning tasks. MSE is primarily used for regression tasks, measuring the average of the squared differences between the predicted and actual values. This makes it advantageous for continuous target variables due to its simplicity and smooth gradient, which benefits gradient-based optimization methods. However, MSE is highly sensitive to outliers because it penalizes larger errors more heavily, and its magnitude depends on the scale of the target variable, which can be problematic for variables with a wide range.
In contrast, Cross-Entropy Loss, also known as Logarithmic Loss, is typically used for classification tasks. It measures the difference between the true and predicted probability distributions. For binary classification, it is defined as the negative average of the sum of the true label times the log of the predicted probability and one minus the true label times the log of one minus the predicted probability. For multi-class classification, it extends to the negative average across all classes. Cross-Entropy Loss provides a probabilistic interpretation of predictions and heavily penalizes confidently incorrect predictions, making it effective for classification problems. It can handle imbalanced datasets well when combined with techniques like class weighting. However, its logarithmic nature can cause numerical instability if predictions are very close to 0 or 1, though this can be mitigated with label smoothing. Also, extremely confident predictions can lead to small gradients, potentially slowing down learning.
Stochastic Gradient Descent (SGD) and its variants are optimization techniques used to update model weights efficiently. Unlike batch gradient descent, which uses the entire dataset, SGD updates weights using a single data point or a small batch at each iteration. This leads to faster convergence, the ability to escape local minima, and introduces update variance. To address the limitations of SGD, several variants have been developed. Momentum accumulates velocity from previous gradients to smooth updates, Adagrad adjusts learning rates based on historical gradient values, and Adam combines elements of both Momentum and Adagrad, offering adaptive learning rates and smoother weight updates. These enhancements make SGD more robust and efficient.
SGD and its variants are particularly useful in scenarios involving large datasets, where computing the gradient over the entire dataset is computationally expensive. Using a single sample or small batch allows for faster updates. In online learning scenarios, where data arrives continuously or is frequently updated, SGD can continuously update the model without retraining on the entire dataset. For non-convex optimization, common in deep learning models, the stochastic nature of SGD helps escape local minima, finding better solutions. Additionally, when the dataset is too large to fit into memory, mini-batch SGD can be used as it requires only a small portion of the data to be loaded at a time.
Implementing MSE, Cross-Entropy Loss, and SGD in nGeneDL Code
In machine learning, selecting the appropriate loss function and optimization technique is crucial for effective model training. Two commonly used loss functions are Mean Squared Error (MSE) and Cross-Entropy Loss, each tailored to different types of tasks. Additionally, Stochastic Gradient Descent (SGD) and its variants offer flexible and efficient ways to optimize model weights. Below, we explore these concepts by quoting specific parts of the provided code, along with their corresponding mathematical equations.
(A) Mean Squared Error (MSE)
Mean Squared Error (MSE) is predominantly used in regression tasks where the goal is to predict continuous target variables. MSE measures the average of the squared differences between the predicted values and the actual values, which helps the model learn by minimizing these differences.
Equation:
MSE = (1/m) * Σ (yi - y_hati)2
In the provided code, MSE is implemented as follows:
if self.task == 'regression':
loss = np.mean((self.a[-1] - y) ** 2) # Mean Squared Error
Here, self.a[-1] represents the model's predictions, and y is the actual target values. The MSE loss function calculates the mean of the squared differences between these two, penalizing larger errors more heavily. This approach is beneficial for tasks requiring continuous output, as it ensures that the model learns to minimize large deviations from the target values.
(B) Cross-Entropy Loss
Cross-Entropy Loss, also known as Logarithmic Loss, is widely used in classification tasks. It measures the difference between the true label distribution and the predicted probability distribution. Cross-Entropy is particularly effective for classification because it penalizes incorrect predictions made with high confidence.
Equation (Binary Classification):
Cross-Entropy = - (1/m) * Σ [yi * log(y_hati) + (1 - yi) * log(1 - y_hati)]
Equation (Multi-Class Classification):
Cross-Entropy = - (1/m) * Σ Σ [yi,c * log(y_hati,c)]
Implementation of Cross-Entropy Loss
The relevant parts of the code for calculating Cross-Entropy Loss are:
if self.task == 'classification':
dz = self.a[-1] - y # Cross-Entropy Loss derivative
and
if self.task == 'classification':
batch_loss = -np.mean(y_batch * np.log(self.a[-1] + 1e-15)) # Cross-Entropy Loss
Breakdown of the Code
Cross-Entropy Loss Derivative:
dz = self.a[-1] - y # Cross-Entropy Loss derivative
self.a[-1]: Represents the predicted probabilities (y_hati for binary classification or y_hati,c for multi-class classification).
y: Represents the true labels, either as binary values (0 or 1) or one-hot encoded vectors for multi-class classification.
dz = self.a[-1] - y: This line calculates the derivative of the Cross-Entropy Loss with respect to the model's output (the predicted probabilities). It reflects how much the predicted probability deviates from the true label.
This derivative is crucial for updating the weights during backpropagation, as it guides the gradient descent process to minimize the loss.
Cross-Entropy Loss Calculation:
if self.task == 'classification':
batch_loss = -np.mean(y_batch * np.log(self.a[-1] + 1e-15)) # Cross-Entropy Loss
Mathematical Translation:
Cross-Entropy = - (1/m) * Σ Σ [yi,c * log(y_hati,c)]
Code Explanation:
np.mean(): Averages the loss over all samples in the batch.
y_batch * np.log(self.a[-1] + 1e-15): This term calculates the log loss for each class and multiplies it by the true label (1 for the correct class, 0 for others). The small value 1e-15 is added to prevent taking the log of zero, which would result in a mathematical error.
Missing Components
The code effectively calculates Cross-Entropy Loss for multi-class classification. However, it does not explicitly handle the second term in the Cross-Entropy formula, which is necessary for binary classification:
- (1 - yi) * log(1 - y_hati)
The current implementation focuses primarily on the term that addresses true positive cases:
yi * log(y_hati)
For multi-class classification, the computation is handled correctly. However, the complementary part (1 - yi), which is relevant for binary classification scenarios, is not explicitly included. Consequently, while the code is appropriate for multi-class tasks, it may not fully account for cases where the model incorrectly predicts a high probability for the negative class in binary classification.
(C) Stochastic Gradient Descent (SGD) and Variants
Stochastic Gradient Descent (SGD) is an optimization technique that updates model weights incrementally using individual data points or small batches, rather than the entire dataset. This approach is computationally efficient and can help the model converge faster while escaping local minima.
Equation (SGD):
θt+1 = θt - η * ∇θ J(θ)
Equation (Momentum):
vt = β1 * vt-1 + η * ∇θ J(θ)
θt+1 = θt - vt
Equation (Adagrad):
Gt = Gt-1 + ∇θ J(θ) ⊙ ∇θ J(θ)
θt+1 = θt - (η / sqrt(Gt + ϵ)) * ∇θ J(θ)
Equation (Adam):
mt = β1 * mt-1 + (1 - β1) * ∇θ J(θ)
vt = β2 * vt-1 + (1 - β2) * (∇θ J(θ))2
m_hatt = mt / (1 - β1t)
v_hatt = vt / (1 - β2t)
θt+1 = θt - (η * m_hatt / (sqrt(v_hatt) + ϵ))
In the code, SGD and its variants are implemented in the backward method:
if self.optimizer == 'SGD':
self.weights[i] -= self.learning_rate * dw # Standard SGD update
elif self.optimizer == 'Momentum':
self.velocity[i] = self.beta1 * self.velocity[i] + self.learning_rate * dw
self.weights[i] -= self.velocity[i] # Momentum update
elif self.optimizer == 'Adagrad':
self.cache[i] += dw ** 2
self.weights[i] -= self.learning_rate * dw / (np.sqrt(self.cache[i]) + self.epsilon) # Adagrad update
elif self.optimizer == 'Adam':
self.t += 1
self.m[i] = self.beta1 * self.m[i] + (1 - the beta1) * dw
self.v[i] = self.beta2 * self.v[i] + (1 - the beta2) * (dw ** 2)
m_hat = self.m[i] / (1 - the beta1 ** self.t)
v_hat = self.v[i] / (1 - the beta2 ** self.t)
self.weights[i] -= self.learning_rate * m_hat / (np.sqrt(v_hat) + self.epsilon) # Adam update
SGD: The standard SGD update adjusts the weights using the gradient (dw) scaled by the learning rate. This method is straightforward and allows for quick updates after each batch.
Momentum: This variant accumulates a velocity term (self.velocity[i]) that smooths the weight updates by considering past gradients, helping to accelerate the convergence.
Adagrad: Adagrad adjusts the learning rate based on the accumulated squared gradients (self.cache[i]). This adaptation helps the model handle sparse data or features by reducing the learning rate for frequently updated weights.
Adam: The Adam optimizer combines the benefits of Momentum and Adagrad by maintaining moving averages of both the gradients (self.m[i]) and the squared gradients (self.v[i]). Adam is particularly effective for deep learning models, offering adaptive learning rates and stability during training.
[Regression] In comparison to the control model (nGeneDL_LinearAlgebra using MSE), the nGeneDL_LinearAlgebra_SGD variants generally perform better with a batch size of 16 than with a batch size of 32, regardless of whether the learning rate is 0.01 or 0.1.
- SGD and its variants (Momentum, Adagrad, Adam) show faster convergence and reduced training times when the batch size is smaller (16), especially noticeable with SGD-Adam, which consistently achieves the quickest training times across different learning rates.
- For larger batch sizes (32), the performance of SGD variants tends to decrease slightly, leading to longer training times and less stable loss values compared to the smaller batch size.
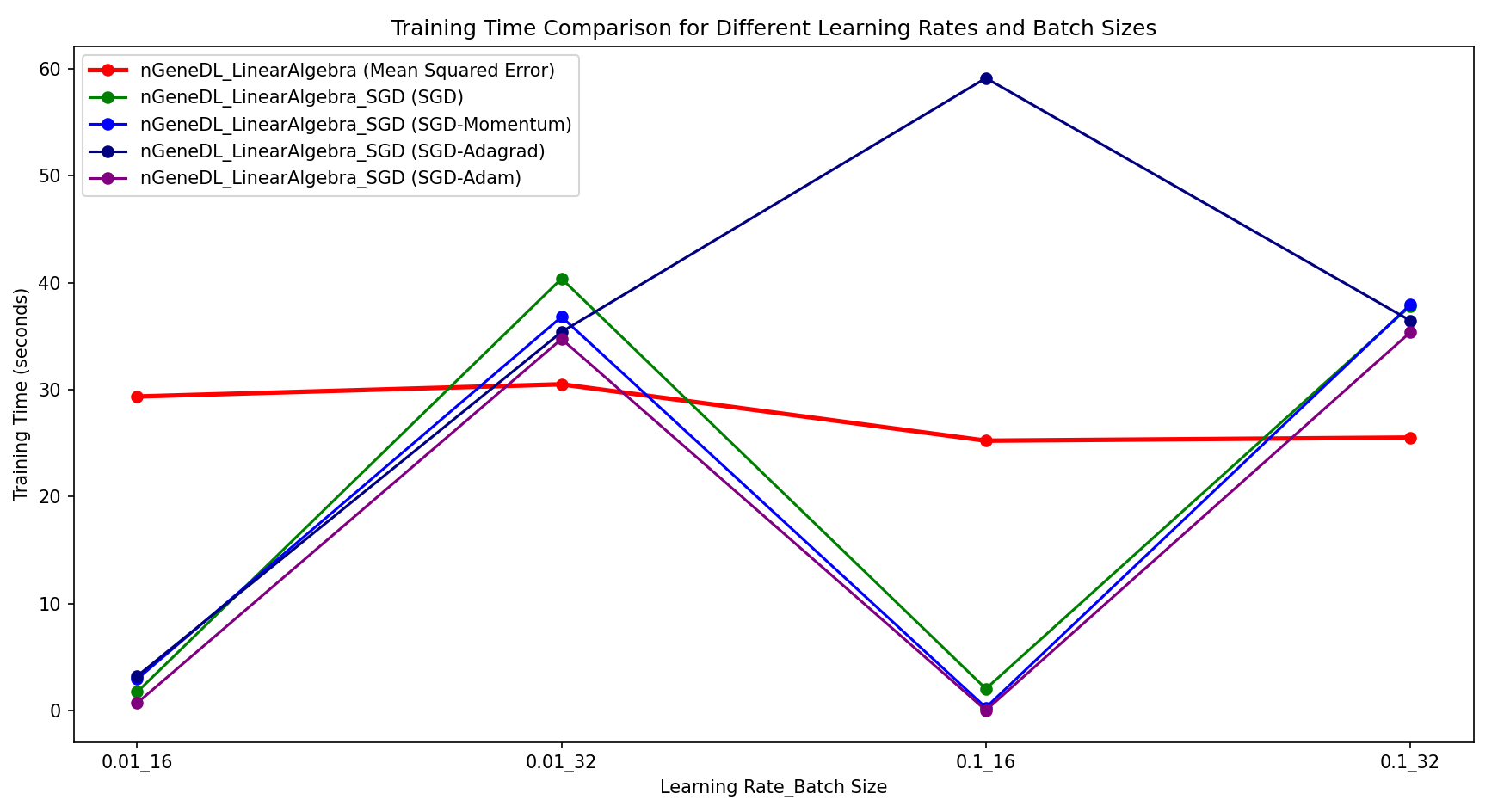
Overall, nGeneDL_LinearAlgebra_SGD demonstrates more efficient training with a batch size of 16, making it preferable for quicker convergence and stability in regression tasks.
[Classification] The results show that for classification tasks, standard SGD of nGeneDL_LinearAlgebra_SGD consistently outperforms both the control model (nGeneDL_LinearAlgebra using Cross-Entropy Loss) and the other SGD variants in all combinations of learning rates and batch sizes.
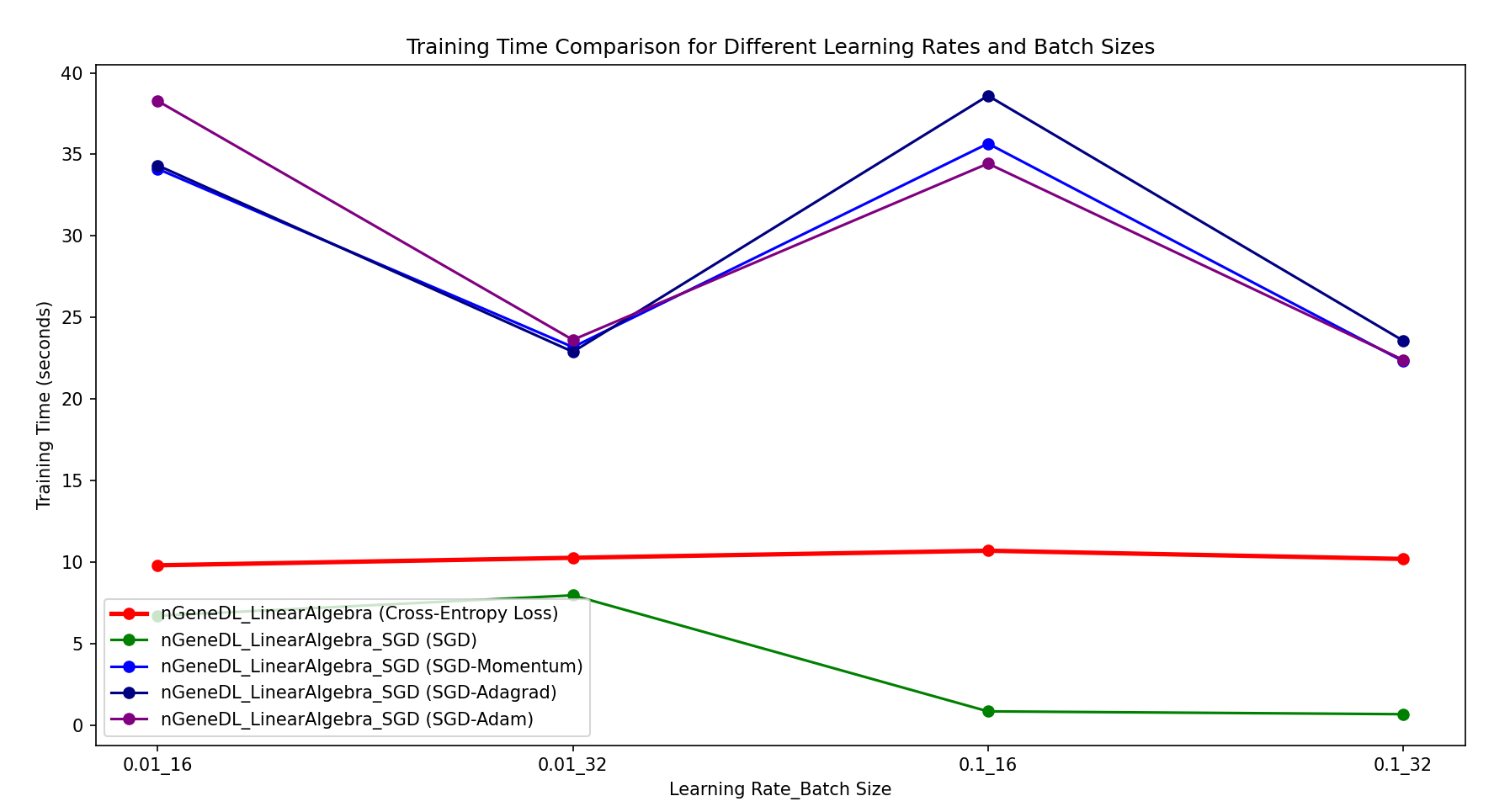
Specifically, standard SGD achieves faster training times and better efficiency compared to both the control model and the other variants (SGD-Momentum, SGD-Adagrad, SGD-Adam), regardless of the learning rate or batch size used.
Convolutional Neural Networks (CNNs)
How Convolutional Neural Networks Work
Convolutional Neural Networks (CNNs) are a class of deep neural networks specifically designed to automatically and adaptively learn spatial hierarchies of features from structured grid data, such as images. These networks are highly effective for image recognition and classification tasks, including applications like handwritten digit recognition. The core components of CNNs include convolutional layers, pooling layers, and fully connected layers, each playing a critical role in the network's functionality.
The convolutional layer is the fundamental building block of a CNN. It consists of a set of learnable filters (or kernels) that slide over the input image, performing convolution operations. These filters detect various features, such as edges, textures, and patterns, by computing the dot product between the filter and local regions of the input. As a result, the convolutional layer generates feature maps that highlight the presence of these features across the image. This process allows the network to learn spatial hierarchies of features, starting from low-level features in the initial layers to high-level, abstract features in deeper layers.
Pooling layers, often inserted between convolutional layers, reduce the spatial dimensions of the feature maps while retaining the most important information. The most common pooling operation is max pooling, which selects the maximum value from each patch of the feature map. Pooling layers help to downsample the data, reducing the computational load and the number of parameters in the network. They also contribute to the invariance of the network to small translations and distortions in the input image, enhancing its robustness.
Finally, fully connected layers, typically located at the end of the CNN, perform high-level reasoning and classification tasks. These layers take the flattened output from the last pooling or convolutional layer and process it through a series of neurons, each connected to every neuron in the previous layer. The fully connected layers integrate the spatially distributed features learned by the convolutional layers and produce the final output, such as class scores in classification tasks.
The combination of convolutional layers, pooling layers, and fully connected layers enables CNNs to effectively learn and recognize complex patterns in images, making them a powerful tool in the field of computer vision.
(A) Weight Sharing and Feature Detection
In a Convolutional Neural Network (CNN), each neuron functions as a visual feature detector by inspecting a small portion of the input image, known as its receptive field. Neurons in the first hidden layer receive specific pixel values as input and produce a high activation if a particular pattern or local visual feature is present within their receptive field.
The function implemented by a neuron is defined by the weights it uses—these weights are represented by the convolutional filter or kernel. When two neurons share the same set of weights but have different receptive fields (i.e., each neuron examines different areas of the input image), they both act as detectors for the same feature but in different locations. This property of sharing the same weights across different neurons is known as weight sharing.
Weight sharing is a fundamental characteristic of CNNs that allows for translation-invariant feature detection. This means that the network can detect the same feature, such as an edge or texture, regardless of where it appears in the image. Because the same filter is applied across the entire image, the network can recognize the feature in any position, making it robust to translations in the input.
(B) Stride Length and Overlapping Receptive Fields
The receptive fields of neurons can overlap, and the amount of overlap is controlled by a hyperparameter called the stride length. For example, if the stride length is one, the receptive field of the neuron is translated by one pixel at each step. Increasing the stride length reduces the overlap between receptive fields.
Stride length refers to how many pixels the filter moves after applying a convolution operation at a given location. Receptive field is the area of the input image that a particular filter covers. When the stride length is small (e.g., 1), the filter moves one pixel at a time, which means there is significant overlap between the receptive fields of consecutive filter applications. Conversely, when the stride length is increased (e.g., to 2 or more), the filter moves more pixels at each step, resulting in less overlap between the receptive fields.
Consider a 3x3 filter applied to a 5x5 image:
Stride Length 1: The filter moves one pixel at a time. Each application of the filter overlaps significantly with the previous one.
Input image (5x5):
[[1, 2, 3, 4, 5],
[6, 7, 8, 9, 10],
[11, 12, 13, 14, 15],
[16, 17, 18, 19, 20],
[21, 22, 23, 24, 25]]
Filter (3x3):
[[1, 2, 3],
[4, 5, 6],
[7, 8, 9]]
Filter application (stride 1):
[[1, 2, 3],
[6, 7, 8],
[11, 12, 13]]
[[2, 3, 4],
[7, 8, 9],
[12, 13, 14]]
... and so on, with overlapping areas
Stride Length 2: The filter moves two pixels at a time. Each application of the filter has less overlap with the previous one compared to stride length 1.
Input image (5x5):
[[1, 2, 3, 4, 5],
[6, 7, 8, 9, 10],
[11, 12, 13, 14, 15],
[16, 17, 18, 19, 20],
[21, 22, 23, 24, 25]]
Filter (3x3):
[[1, 2, 3],
[4, 5, 6],
[7, 8, 9]]
Filter application (stride 2):
[[1, 2, 3],
[6, 7, 8],
[11, 12, 13]]
[[3, 4, 5],
[8, 9, 10],
[13, 14, 15]]
... and so on, with less overlap
In summary, increasing the stride length reduces the overlap between receptive fields, allowing the filter to cover more unique regions of the input image. This reduction in redundancy can make the convolution operations more efficient, although it may also lead to a loss of some spatial detail.
(C) Convolution Operation
In computer vision, the matrix of weights applied to an input is known as the kernel (or convolution mask). The operation of sequentially passing a kernel across an image and within each local region, weighting each input and adding the result to its local neighbors, is known as a convolution.
Convolving a kernel across an image is equivalent to passing a local visual feature detector across the image and recording all the locations in the image where the visual feature was present. The output from this process is a map of all the locations in the image where the relevant visual feature occurred. For this reason, the output of a convolution process is sometimes known as a feature map.
A kernel, also known as a convolution mask or filter, is a small matrix of weights used in the context of images. Typically smaller than the input image, the kernel scans over the image to detect specific features. Convolution is the process of sliding the kernel across the image and performing element-wise multiplication and summation to produce a feature map. This operation helps in identifying and extracting various features from the input image.
The process of convolution begins with sliding the kernel across the input image. The stride, which is the number of pixels the kernel moves at each step, determines the movement. At each position, a sub-section of the image, equal in size to the kernel, is selected. Next, element-wise multiplication is performed between the kernel and the selected sub-section of the image. The results of these multiplications are then summed to produce a single value, which becomes an element in the output feature map. The kernel then moves to the next position based on the stride and repeats this process until it has scanned the entire image. By continuously moving and applying the kernel across the image, the convolution operation extracts and highlights specific features, ultimately forming a detailed feature map.
Let's consider a simple example with a 3x3 kernel and a 5x5 image.
Input Image (5x5):
1 2 3 4 5
6 7 8 9 10
11 12 13 14 15
16 17 18 19 20
21 22 23 24 25
Kernel (3x3):
1 0 -1
1 0 -1
1 0 -1
Convolution Steps
(1) First Position (Top-left corner): Select the top-left 3x3 sub-section of the image. Multiply element-wise with the kernel and sum the results.
Sub-section of the image:
1 2 3
6 7 8
11 12 13
Kernel:
1 0 -1
1 0 -1
1 0 -1
Element-wise multiplication:
1*1 + 2*0 + 3*(-1) + 6*1 + 7*0 + 8*(-1) + 11*1 + 12*0 + 13*(-1)
= 1 + 0 - 3 + 6 + 0 - 8 + 11 + 0 - 13
= -6
Output value at (1,1): -6
(2) Next Position (Move kernel to the right by the stride, here stride is 1): Repeat the process for the next 3x3 sub-section. Continue this process until the kernel has scanned the entire image.
Resulting Feature Map
After the kernel has been applied to the entire image, the resulting feature map (or output) could look like this (assuming stride 1 and valid padding):
-6 -6 -6 -6
-6 -6 -6 -6
-6 -6 -6 -6
-6 -6 -6 -6
A kernel, also known as a convolution mask or filter, is a small matrix of weights used to scan over an image. The process of convolution involves sliding this kernel across the image and performing element-wise multiplication and summation to produce a feature map. This feature map is the output of the convolution process and highlights the locations in the image where specific features are detected. By detecting features such as edges, corners, and textures, this process provides a clearer understanding of how convolution operations work in computer vision and their role in Convolutional Neural Networks (CNNs).
Relevant Code:
def conv2d(self, X, filter_size, stride):
...
for i in range(0, input_height - filter_height + 1, stride):
for j in range(0, input_width - filter_width + 1, stride):
region = X[i:i + filter_height, j:j + filter_width]
output[i // stride, j // stride] = np.sum(region)
- Weight Sharing: In your
conv2dmethod, the same filter is applied to different regions of the input image as the nested loops iterate over it. This means the same weights are used across the image, ensuring that the feature is detected across different parts of the image. - Feature Detection: The line
output[i // stride, j // stride] = np.sum(region)computes the activation for each receptive field by summing up the values within the region, mimicking the detection of a specific feature represented by the filter. - Stride Length: The
strideparameter in the loops controls how many pixels the filter moves after each application. A smaller stride results in more overlap between consecutive receptive fields, whereas a larger stride reduces this overlap. In your code, the stride is set by theself.strideattribute when callingconv2d. - Convolution Operation: The
conv2dfunction implements the convolution operation by taking a 2D inputX, and applying a filter of sizefilter_sizeacross it. The nested loops iterate over the image, selecting sub-regions (region) of the image, and computing the sum of the element-wise multiplication, which is stored in theoutputmatrix.
(A) Weight Sharing and Feature Detection
(B) Stride Length and Overlapping Receptive Fields
(C) Convolution Operation
(D) Activation Functions
The convolution operation in Convolutional Neural Networks (CNNs) does not inherently include a nonlinear activation function; it primarily involves a weighted summation of the inputs to identify basic patterns in the data, such as edges and textures. To capture more complex patterns, it is standard to apply a nonlinearity operation to the feature maps generated by convolutional layers. This is often accomplished using the Rectified Linear Unit (ReLU) activation function, defined as: rectifier(z) = max(0, z). The ReLU function introduces nonlinearity by transforming each position in the feature map, setting all negative values to zero while retaining positive values. This transformation allows the network to learn intricate patterns and relationships that linear operations alone cannot capture.
After convolution and ReLU activation, the next critical step is dimensionality reduction, achieved through pooling layers. Pooling layers, such as max pooling, are employed to condense the features extracted by the convolutional layers, reducing the spatial dimensions of the data. This process retains only the most important aspects of the feature maps, leading to a more manageable and robust representation of the input. Pooling helps the network become less sensitive to variations such as translations or noise, thereby enhancing its ability to generalize from the input data.
The typical sequence of operations in CNNs, namely Convolution → ReLU Activation → Max Pooling, works synergistically to extract meaningful features, introduce nonlinearity, and efficiently down-sample the data, creating a powerful framework for tasks like image recognition and classification.
def relu(self, X):
return np.maximum(0, X)
- ReLU Activation: The
relufunction is applied to the output of the convolutional layer to introduce non-linearity. This function replaces all negative values in the feature map with zero, which allows the network to learn complex patterns by stacking multiple layers.
(E) Pooling
Pooling operations in Convolutional Neural Networks (CNNs) are crucial techniques used to reduce the spatial dimensions of input feature maps while preserving important information. Like convolution operations, pooling involves repeatedly applying a function across an input space, but its primary goal is to down-sample the feature maps, decreasing computational complexity and mitigating overfitting by making the representation smaller and more manageable.
There are two main types of pooling used in CNNs: max pooling and average pooling. Max pooling selects the maximum value from a defined region, or pooling window, effectively capturing the most prominent feature in that region. This makes max pooling particularly effective for retaining sharp, high-intensity features. In contrast, average pooling calculates the average value of the inputs within the pooling window, providing a more generalized representation of the features and smoothing out noise. Both techniques operate over a small region of the input feature map, known as the receptive field.
The size of the receptive field determines the region over which the pooling function operates, and the stride determines how far the pooling window moves after each operation. If the stride is greater than one, the pooling windows do not overlap, resulting in a more significant reduction in spatial dimensions. This non-overlapping nature ensures distinct regions are sampled, whereas overlapping windows may capture more detail but require more computational resources.
In CNNs, the typical sequence of operations involves applying convolutional filters to the input image to generate feature maps, followed by the application of non-linear activation functions like ReLU (Rectified Linear Unit) to introduce nonlinearity. This is then followed by pooling, which helps down-sample the feature maps. By reducing the height and width of these maps, pooling helps the network become invariant to small translations and distortions in the input image, significantly lowering the number of parameters and operations required. This makes the network more efficient, enabling faster training and inference, which is especially beneficial for large-scale and real-time applications.
A-5-1) Max Pooling: Let's say we have a 4x4 input feature map and a 2x2 pooling window with a stride of 2 (no overlap).
Input Feature Map (4x4):
1 3 2 4
5 6 1 2
9 8 7 6
4 3 2 1
Max Pooling (2x2 window, stride 2):
First Position (Top-left 2x2 region), with maximum value of 6
1 3
5 6
Second Position (Top-right 2x2 region), with maximum value of 4
2 4
1 2
Third Position (Bottom-left 2x2 region), with maximum value of 9
9 8
4 3
Fourth Position (Bottom-right 2x2 region), with maximum value of 7
7 6
2 1
Resulting Feature Map after Max Pooling:
6 4
9 7
A-5-2) Average Pooling: Let's apply the same 2x2 pooling window with a stride of 2 to perform average pooling.
First Position (Top-left 2x2 region), with average value of (1+3+5+6)/4 = 3.75
1 3
5 6
Second Position (Top-right 2x2 region), with average value of (2+4+1+2)/4 = 2.25
2 4
1 2
Third Position (Bottom-left 2x2 region), with average value of (9+8+4+3)/4 = 6
9 8
4 3
Fourth Position (Bottom-right 2x2 region), with average value of (7+6+2+1)/4 = 4
7 6
2 1
Resulting Feature Map after Average Pooling:
3.75 2.25
6.00 4.00
def max_pooling(self, X, pool_divisions):
input_height, input_width = X.shape
pool_size = input_height // pool_divisions
output = np.zeros((pool_divisions, pool_divisions))
for i in range(pool_divisions):
for j in range(pool_divisions):
region = X[i * pool_size:(i + 1) * pool_size, j * pool_size:(j + 1) * pool_size]
output[i, j] = np.max(region)
return output
- Max Pooling: The
max_poolingmethod performs max pooling by dividing the feature map into non-overlapping regions based on thepool_divisionsparameter. For each region, it takes the maximum value, effectively down-sampling the feature map and retaining the most significant features. - Reduction in Spatial Dimensions: By selecting the maximum value from each pooling region, this function reduces the spatial size of the feature map, which helps in reducing computational load and makes the network more robust.
(F) Multiple Convolutional Layers and Filters
A CNN can generalize beyond one feature by training multiple convolutional layers in parallel (or filters), with each filter learning a single kernel matrix (feature detection function). The outputs of multiple filters can be integrated in a variety of ways. One way is to combine the feature maps generated by separate filters into a single multifilter feature map. A subsequent convolutional layer then takes this multifilter feature map as input.
The nGeneCNN_MultipleFilters Class: The nGeneCNN_MultipleFilters class extends the functionality of the nGeneCNN_Prototype class to support multiple convolutional filters. This enhancement aligns with the concept of having multiple convolutional layers and filters in a CNN, which allows the network to generalize beyond detecting a single feature and instead detect multiple distinct features within the input data.
1. Initialization of Multiple Filters
def __init__(self, filter_size=(3, 3), stride=2, pool_divisions=-1, num_filters=3, percentage_combinations=100, debug_backpropagation=False):
super().__init__(filter_size=filter_size, stride=stride, pool_divisions=pool_divisions, debug_backpropagation=debug_backpropagation)
self.num_filters = num_filters
self.percentage_combinations = percentage_combinations
self.filters = self.initialize_filters()
- The
__init__method initializes multiple filters for the convolutional layer. The number of filters is determined by thenum_filtersparameter. initialize_filtersgenerates these filters randomly, which allows each filter to potentially learn a different feature during the training process.
2. Filter Initialization
def initialize_filters(self):
total_combinations = 2 ** (self.filter_size[0] * self.filter_size[1])
num_filters_to_generate = int((self.percentage_combinations / 100.0) * total_combinations)
filters = np.random.randn(min(self.num_filters, num_filters_to_generate), self.filter_size[0], self.filter_size[1])
return filters
- The
initialize_filtersmethod generates a set of filters based on the specifiedfilter_size,num_filters, andpercentage_combinations. - This ensures that each filter is unique and can detect different features within the input data, allowing the network to capture a broader range of patterns.
3. Convolution with Multiple Filters
def conv2d(self, X, filter_size, stride):
filter_height, filter_width = filter_size
input_height, input_width = X.shape
output_height = (input_height - filter_height) // stride + 1
output_width = (input_width - filter_width) // stride + 1
output = np.zeros((self.num_filters, output_height, output_width))
for f in range(self.num_filters):
for i in range(0, input_height - filter_height + 1, stride):
for j in range(0, input_width - filter_width + 1, stride):
region = X[i : i + filter_height, j : j + filter_width]
output[f, i // stride, j // stride] = np.sum(region * self.filters[f])
return output- The
conv2dmethod has been modified to perform convolution operations using multiple filters. - For each filter, the method iterates over the input data and applies the filter to different regions of the input.
- The result is a set of feature maps, one for each filter, which are stored in a 3D output array (where each "slice" along the first dimension corresponds to the feature map from one filter).
4. Max Pooling Across Multiple Feature Maps
def max_pooling(self, X, pool_divisions):
num_filters, input_height, input_width = X.shape
pool_size = input_height // pool_divisions
output = np.zeros((num_filters, pool_divisions, pool_divisions))
for f in range(num_filters):
for i in range(pool_divisions):
for j in range(pool_divisions):
region = X[f, i * pool_size : (i + 1) * pool_size, j * pool_size : (j + 1) * pool_size]
output[f, i, j] = np.max(region)
return output- The
max_poolingmethod applies max pooling independently to each feature map generated by the filters. - This method processes each feature map separately, down-sampling it by taking the maximum value within each pooling region.
- This reduces the spatial dimensions of each feature map while retaining the most important information, thereby making the feature maps more manageable for further processing.
5. Visualization of Filters and Feature Maps
def visualize_filters(self):
num_filters = len(self.filters)
fig, axes = plt.subplots(1, num_filters, figsize=(15, 5))
for i, ax in enumerate(axes):
ax.imshow(self.filters[i], cmap='gray')
ax.set_title(f'Filter {i+1}')
ax.axis('off')
plt.show()def visualize_feature_maps(self, feature_maps, title="Feature Maps"):
num_feature_maps = feature_maps.shape[0]
fig, axes = plt.subplots(1, num_feature_maps, figsize=(15, 5))
for i, ax in enumerate(axes):
ax.imshow(feature_maps[i], cmap='gray')
ax.set_title(f'Feature Map {i+1}')
ax.axis('off')
plt.suptitle(title)
plt.show()visualize_filtersallows for the visualization of each filter, making it possible to inspect what each filter has learned.visualize_feature_mapsenables the visualization of the resulting feature maps after applying the filters and pooling, helping to understand which features are being detected by the network.
Example: Handwritten Digit Recognition
In the context of handwritten digit recognition, a CNN might receive an input image of a digit (e.g., 28x28 pixels). The first layer could apply several convolutional filters to detect edges and simple shapes. Subsequent layers would detect more complex features, such as curves and digit-like shapes, by combining the features detected in previous layers. Pooling layers would reduce the spatial dimensions, preserving the most important information. Finally, dense layers would integrate these features to classify the image as one of the ten possible digits (0-9).
By leveraging weight sharing, convolutional operations, activation functions, and pooling, CNNs efficiently and effectively recognize patterns in images, making them powerful tools for tasks like handwritten digit recognition.
Overview of Training Approaches
This analysis explores three primary training methods on the MNIST digit recognition task:
- No CNN, Cross Entropy Loss Optimization: Utilizes a fully connected neural network trained with Cross Entropy Loss, without any convolutional layers.
- No CNN, SGD Optimization: Similar to the first method but uses Stochastic Gradient Descent (SGD) and its variants as the optimization method.
- CNN with Single Filter, Comparison of Time & Accuracy: Introduces a single convolutional filter and compares performance across different optimizers, focusing on both training time and accuracy.
(A) No CNN with Cross Entropy Loss Optimization
In the train_and_test_mnist_no_cnn_nGeneDL_LinearAlgebra function, a fully connected neural network is trained on the MNIST dataset using Cross Entropy Loss as the loss function. This method does not utilize any convolutional layers, and the network relies solely on dense layers to learn the digit patterns.
=== [Digit Recognition] no CNN by Cross Entropy Loss Optimization === Epoch 1/1000: Loss = 0.2454645 Epoch 201/1000: Loss = 0.0386511 Epoch 401/1000: Loss = 0.0301550 Epoch 601/1000: Loss = 0.0268234 Epoch 801/1000: Loss = 0.0242453 => Test set accuracy: 0.9287
- Training: The network is trained over 1000 epochs, with threshold-based early stopping if the loss falls below a certain threshold.
- Performance: While this method achieves a test accuracy of approximately 92.87%, it takes significantly longer to converge, as indicated by the gradual decrease in loss over the epochs.
nn = nGeneDL_LinearAlgebra(input_nodes=784, output_nodes=10, hidden_nodes=[128, 64], learning_rate=0.1)
nn.fit(X_train.T, Y_train_one_hot, epochs=1000, threshold_based_early_stopping=True)
test_predictions = nn.predict(X_test.T)
accuracy = np.sum(test_predictions.flatten() == Y_test) / Y_test.size
print(f"=> Test set accuracy: {accuracy}")
- Epochs: 1000 (with early stopping).
- Final Loss: 0.0242453.
- Test Accuracy: 0.9287.
(B) No CNN with SGD Optimization
The train_and_test_mnist_no_cnn_nGeneDL_LinearAlgebra_SGD function also trains a fully connected network but uses various SGD-based optimizers, including Momentum, Adagrad, and Adam. This method improves both convergence speed and accuracy compared to Cross Entropy Loss optimization.
=== [Digit Recognition] no CNN by SGD Optimization with Single Filter ===
Training with optimizer: SGD, learning rate: 0.01, batch size: 32
Epoch 1/1000: Loss = 0.1041797
-> Threshold-based early stopping at epoch 28 with loss 0.0041823 as loss < 0.005.
===> Test set accuracy: 0.9257
Training with optimizer: Momentum, learning rate: 0.01, batch size: 32
Epoch 1/1000: Loss = 0.0766008
-> Threshold-based early stopping at epoch 14 with loss 0.0010442 as loss < 0.005.
===> Test set accuracy: 0.9556
Training with optimizer: Adagrad, learning rate: 0.01, batch size: 32
Epoch 1/1000: Loss = 0.0209173
-> Threshold-based early stopping at epoch 4 with loss 0.0042766 as loss < 0.005.
===> Test set accuracy: 0.9350
Training with optimizer: Adam, learning rate: 0.01, batch size: 32
Epoch 1/1000: Loss = 0.0443852
-> Threshold-based early stopping at epoch 3 with loss 0.0041846 as loss < 0.005.
===> Test set accuracy: 0.9444
- Training: The network is trained using different optimizers, which adjust the learning rate dynamically or incorporate momentum to accelerate convergence.
- Performance: With optimizers like Momentum and Adam, the network achieves higher accuracy (up to 95.56%) and converges much faster, often within a few epochs.
nn_sgd = nGeneDL_LinearAlgebra_SGD(input_nodes=784, output_nodes=10, hidden_nodes=[128, 64], learning_rate=0.01, optimizer='Adam')
nn_sgd.fit(X_train.T, Y_train_one_hot, epochs=1000, batch_size=32, early_stopping=False)
test_predictions = nn_sgd.predict(X_test.T)
accuracy = np.sum(test_predictions.flatten() == Y_test) / Y_test.size
print(f"===> Test set accuracy: {accuracy:.4f}")- Best Optimizer: Momentum (Accuracy: 0.9556).
- Fastest Convergence: Adam (Converged within 2-4 epochs).
- Test Accuracy Range: 0.9257 to 0.9556.
(C) CNN with Single Filter: Time & Accuracy Comparison
The train_and_test_mnist_cnn_SGD_comparision_time_accuracy_without_prediction function introduces a CNN architecture with a single convolutional filter. The function compares the performance of the Cross Entropy Loss and various SGD optimizers in terms of both training time and accuracy.
=== [Digit Recognition] CNN by SGD Optimizations with Single Filter for Comparison: Time & Accuracy === Original raw input dimensions: (28, 28) Feature map dimension after convolution: (26, 26) Possible non-overlapping pool divisions: [1, 2, 13] => Maximum non-overlapping pool divisions possible (excluding feature map dimension): 13 Processing image for X_train_processed:.......... Processing image for X_test_processed :.......... => Number of Input nodes: 169 === Training Time & Prediction Accuracy === Epoch 1/1000: Loss = 0.5406638 Epoch 201/1000: Loss = 0.0386366 Epoch 401/1000: Loss = 0.0309034 Epoch 601/1000: Loss = 0.0270548 Epoch 801/1000: Loss = 0.0244385 ===> nGeneDL_LinearAlgebra (Cross Entropy Loss) training time: 73.79 seconds ===> Test set accuracy for nGeneDL_LinearAlgebra (Cross Entropy Loss): 0.9304 Epoch 1/1000: Loss = 0.0581560 -> Threshold-based early stopping at epoch 2 with loss 0.0046510 as loss < 0.005. ===> nGeneDL_LinearAlgebra_SGD (SGD) training time: 0.22 seconds ===> Test set accuracy for nGeneDL_LinearAlgebra_SGD (SGD): 0.9018 Epoch 1/1000: Loss = 0.0415230 -> Threshold-based early stopping at epoch 7 with loss 0.0020228 as loss < 0.005. ===> nGeneDL_LinearAlgebra_SGD (Momentum) training time: 0.80 seconds ===> Test set accuracy for nGeneDL_LinearAlgebra_SGD (Momentum): 0.9642 Epoch 1/1000: Loss = 0.0135918 -> Threshold-based early stopping at epoch 4 with loss 0.0047833 as loss < 0.005. ===> nGeneDL_LinearAlgebra_SGD (Adagrad) training time: 0.54 seconds ===> Test set accuracy for nGeneDL_LinearAlgebra_SGD (Adagrad): 0.9524 Epoch 1/1000: Loss = 0.0191139 -> Threshold-based early stopping at epoch 2 with loss 0.0042699 as loss < 0.005. ===> nGeneDL_LinearAlgebra_SGD (Adam) training time: 0.28 seconds ===> Test set accuracy for nGeneDL_LinearAlgebra_SGD (Adam): 0.9103
- CNN Architecture: A single convolutional filter is applied, followed by max pooling. The processed data is then passed to a fully connected network.
- Training Efficiency: This method significantly reduces training time while maintaining high accuracy, especially when using optimizers like Momentum and Adam.
preprocessor = nGeneCNN_Prototype(filter_size=(3, 3), stride=1)
X_train_processed = preprocessor.preprocess(X_train)
X_test_processed = preprocessor.preprocess(X_test)
nn = nGeneDL_LinearAlgebra(input_nodes=X_train_processed.shape[1], output_nodes=10, hidden_nodes=[128, 128], learning_rate=0.01)
nn.fit(X_train_processed, Y_train_one_hot, epochs=1000, threshold_based_early_stopping=True)
test_predictions = nn.predict(X_test_processed)
accuracy = np.sum(test_predictions.flatten() == Y_test) / Y_test.size
print(f"===> Test set accuracy: {accuracy}\n")- Cross Entropy Loss Training Time: 73.79 seconds.
- Best Optimizer: Momentum (Accuracy: 0.9642, Training Time: 0.80 seconds).
- Test Accuracy Range: 0.9018 to 0.9642.
(D) Analysis of Digit Recognition Using CNN with Multiple Filters
This analysis examines the application of Convolutional Neural Networks (CNNs) with three filters to the MNIST digit recognition task. The objective is to compare the performance of two frameworks: nGeneDL_LinearAlgebra using Cross Entropy Loss, and nGeneDL_LinearAlgebra_SGD using Stochastic Gradient Descent (SGD) and its variants (Momentum, Adagrad, and Adam). The focus is on evaluating each method in terms of both training efficiency and accuracy.
Multiple Filters and Feature Visualization: Prior to discussing the results, the model visualizes the initial random filters and their processing of the hand-digit images. This visualization is crucial for understanding how each filter detects different aspects of the digits, such as edges, textures, or shapes.
- Filter Visualization: The three filters begin with random weights, which evolve as the network undergoes training. Each filter is designed to focus on different features within the image, contributing to a more comprehensive understanding of the data.
- Feature Map Visualization: Following the convolution operation, the resulting feature maps highlight the areas of the image that each filter finds significant, providing insights into the learning process.


D-1) nGeneDL_LinearAlgebra with Cross Entropy Loss
The train_and_test_mnist_cnn_multiple_filters function tests the CNN with multiple filters using the nGeneDL_LinearAlgebra framework and Cross Entropy Loss. The model is trained for a full 1000 epochs, as the loss does not drop below 0.005 before reaching the later epochs.
- Training Duration: The network completes all 1000 epochs because the reduction in loss is gradual, requiring the entire training period to achieve the desired level.
- Test Accuracy: The model achieves a test set accuracy of 92.84%, demonstrating strong performance, albeit with longer training times.
- Loss Trajectory: The loss decreases steadily over time, indicating that the model is learning effectively, though at a slower pace compared to other optimizers.
Epoch 1/1000: Loss = 0.3411547 Epoch 201/1000: Loss = 0.0410656 Epoch 401/1000: Loss = 0.0298363 Epoch 601/1000: Loss = 0.0254221 Epoch 801/1000: Loss = 0.0229610 => Test set accuracy: 0.9284
D-2) nGeneDL_LinearAlgebra_SGD with SGD or its variants
The train_and_test_mnist_cnn_SGD_multiple_filters function evaluates the CNN with multiple filters using the nGeneDL_LinearAlgebra_SGD framework with various SGD-based optimizers. The results demonstrate that these optimizers significantly reduce the training time by triggering threshold-based early stopping when the loss drops below 0.005.
a. SGD Optimizer
- Training Duration: The model stops at epoch 15, as the loss quickly falls below the threshold.
- Test Accuracy: The test accuracy reaches 93.63%, indicating a good balance between training speed and model performance.
Epoch 1/1000: Loss = 0.0986447 -> Threshold-based early stopping at epoch 15 with loss 0.0048741 as loss < 0.005. ===> Test set accuracy for nGeneDL_LinearAlgebra_SGD (SGD): 0.9363
b. Momentum Optimizer
- Training Duration: The fastest optimizer in this comparison, stopping at epoch 5 with a loss of 0.0035961.
- Test Accuracy: Momentum achieves the highest accuracy at 96.21%, demonstrating its effectiveness in quickly converging to an optimal solution.
Epoch 1/1000: Loss = 0.0264336 -> Threshold-based early stopping at epoch 5 with loss 0.0035961 as loss < 0.005. ===> Test set accuracy for nGeneDL_LinearAlgebra_SGD (Momentum): 0.9621
c. Adagrad Optimizer
- Training Duration: Similar to Momentum, Adagrad stops early at epoch 5, but with a slightly higher loss of 0.0040192.
- Test Accuracy: Adagrad achieves a strong test accuracy of 95.87%, making it another viable option for this task.
Epoch 1/1000: Loss = 0.0594092 -> Threshold-based early stopping at epoch 5 with loss 0.0040192 as loss < 0.005. ===> Test set accuracy for nGeneDL_LinearAlgebra_SGD (Adagrad): 0.9587
d. Adam Optimizer
- Training Duration: Adam converges the fastest, stopping at epoch 3 with a loss of 0.0022884.
- Test Accuracy: While not the highest, Adam still achieves a robust accuracy of 94.85%, highlighting its efficiency in minimizing loss quickly.
Epoch 1/1000: Loss = 0.0545857 -> Threshold-based early stopping at epoch 3 with loss 0.0022884 as loss < 0.005. ===> Test set accuracy for nGeneDL_LinearAlgebra_SGD (Adam): 0.9485
Conclusion
- Training Efficiency:
nGeneDL_LinearAlgebra_SGDwith SGD or its variants consistently finishes earlier thannGeneDL_LinearAlgebrawith Cross Entropy Loss. Among the SGD variants, Momentum emerges as the best in terms of both speed and accuracy, followed closely by Adagrad and Adam. - Test Accuracy: While all methods achieve strong test accuracy, Momentum stands out with the highest accuracy of 96.21%.
- Recommendation: For browser-based or time-sensitive applications,
nGeneDL_LinearAlgebra_SGDwith Momentum or Adam is recommended due to their quick convergence and high accuracy.
By leveraging multiple filters and the appropriate optimizer, the model efficiently processes digit images, leading to faster training times and better generalization on test data.
Reference (listed in order of greatest to least influence)
- Kelleher, J. D. (2019). Deep Learning. MIT Press. ISBN: 9780262537551.
- Starmer, J. (n.d.; 2022). StatQuest with Josh Starmer [YouTube channel] and The StatQuest Illustrated Guide To Machine Learning. StatQuest Publications. ISBN-13: 979-8986924007. Retrieved from https://www.youtube.com/watch?v=zxagGtF9MeU&list=PLblh5JKOoLUIxGDQs4LFFD--41Vzf-ME1
- Zhang, S. (n.d.). Simple MNIST NN from scratch (numpy, no TF/Keras) [Video and source code]. YouTube; Kaggle. Retrieved from https://www.youtube.com/watch?v=w8yWXqWQYmU&t=13s and https://www.kaggle.com/code/wwsalmon/simple-mnist-nn-from-scratch-numpy-no-tf-keras
- Knowings, L. D. (2024). Building neural networks from scratch with Python. Independently published.
- Géron, A. (2022). Hands-On Machine Learning with Scikit-Learn, Keras, and TensorFlow: Concepts, Tools, and Techniques to Build Intelligent Systems (3rd ed.). O'Reilly Media.
- Raschka, S., & Mirjalili, V. (2019). Python Machine Learning: Machine Learning and Deep Learning with Python, scikit-learn, and TensorFlow 2 (3rd ed.). Packt Publishing.
- Raschka, S., & Liu, Y. (2022). Machine Learning with PyTorch and Scikit-Learn: Develop machine learning and deep learning models with Python. Packt Publishing.
- NeuralNine. (n.d.). NeuralNine [YouTube channel]. Retrieved from https://www.youtube.com/@NeuralNine